Nicholas Owen
Research Fellow and Bioinformatician
University College London
The Francis Crick Institute
Moorfields Eye Hospital NHS
Biography
I am a senior postdoctoral research fellow at the UCL Insititute of Ophthalmology in the Ocular Genomics and Therapeutics group of Professor Mariya Moosajee.
My research focuses on the molecular and cellular biology of rare diseases. Investigating the biological mechanisms involved in the genetic disease Spinal Muscular Atrophy (SMA), I developed an interest in RNA splicing and disease. My work developed and optimised an antisense based method to modulate the alternative splicing pattern of transcripts as a therapy for patients.
Since then, I have been engaged in a several research projects involving RNA splicing defects, including retinitis pigmentosa (RP) and Alzheimer’s Disease. I have continued my interests in improving our understanding of rare diseases through transcriptomic (RNA-seq), epigenetic and whole genome data analysis. With a honorary research position at Moorfields Eye Hospital I have been fortunate to investigate several genetic ocular disorders and utilise the wealth of clinical information to enhance the analysis of patient transcriptome and whole genome sequence data.
Download my resumé .
- Rare disease research
- RNA, splicing and disease
- Bioinformatics
- Transcriptomics & Genomics
- Personalised Medicine and Public Health
- Public and Patient Engagement
-
DPhil in Molecular Genetics
Oxford University
-
M.Sc. Human Molecular Genetics
St. Marys Hospital/Imperial College
-
B.Sc (Hons) Genetics
QMUL, London
Skills
R for Statistics
Human Disease Modelling
Open Code and Data
Cell & Molecular Methods
Reproducible Code
Mentoring/Training
Science Communication
Version Control
HDR, Astronomy
Open Science Framework
Research Projects
Recent & Upcoming Talks
Recent Posts
Featured Publications

Ocular coloboma (OC) is a failure of complete optic fissure closure during embryonic development and presents as a tissue defect along the proximal distal axis of the ventral eye. It is classed as part of the clinical spectrum of structural eye malformations with microphthalmia and anophthalmia, collectively abbreviated to MAC. Despite deliberate attempts to identify causative variants in MAC, many patients remain without a genetic diagnosis. To reveal potential candidate genes, we utilised transcriptomes experimentally generated from embryonic eye tissues derived from human, mouse, zebrafish, and chicken at stages coincident with optic fissure closure
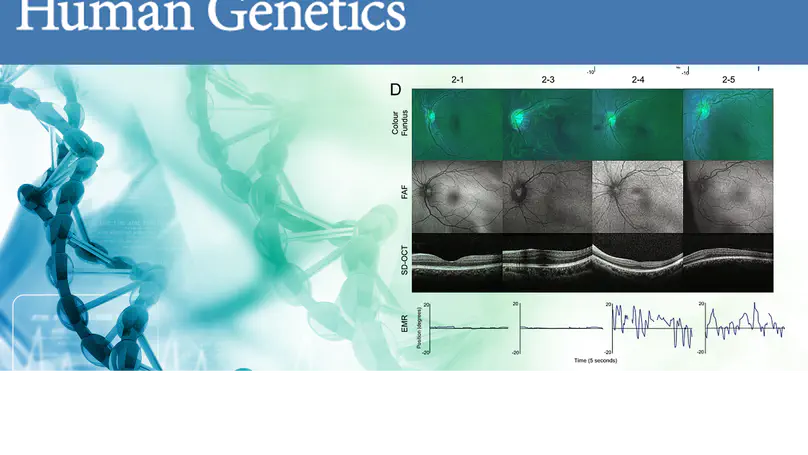
The crumbs cell polarity complex plays a crucial role in apical-basal epithelial polarity, cellular adhesion, and morphogenesis. Homozygous variants in human CRB1 result in autosomal recessive Leber congenital amaurosis (LCA) and retinitis pigmentosa (RP), with no established genotype-phenotype correlation. We performed integrative transcriptomic and methylomic analysis of whole retina to identify dysregulated genes and pathways.

Purpose: Ocular coloboma arises from genetic or environmental perturbations that inhibit optic fissure (OF) fusion during early eye development. Despite high genetic heterogeneity, 70% to 85% of patients remain molecularly undiagnosed. In this study, we have identified new potential causative genes using cross-species comparative meta-analysis.
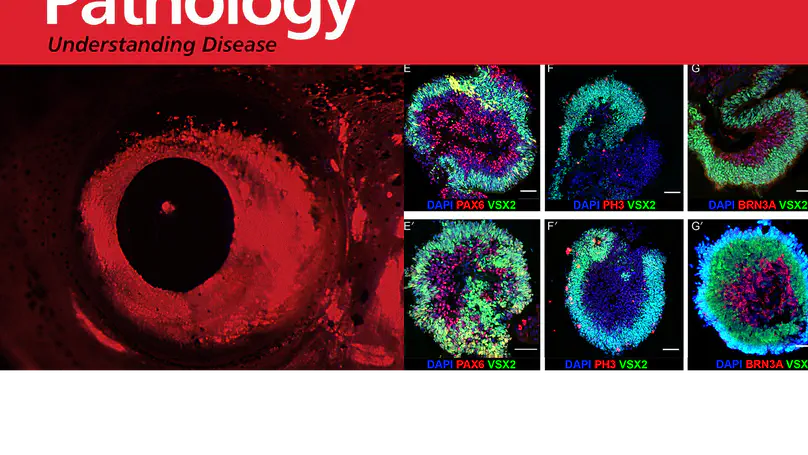
PAX6 is considered the master regulator of eye development, the majority of variants affecting this gene cause the pan-ocular developmental eye disorder aniridia. Although no genotype-phenotype correlations are clearly established, missense variants affecting the DNA-binding paired domain of PAX6 are usually associated with non-aniridia phenotypes like microphthalmia, coloboma or isolated foveal hypoplasia. In this study, we report …

High-throughput, massively parallel sequence analysis has revolutionized the way that researchers design and execute scientific investigations. Vast amounts of sequence data can be generated in short periods of time. Regarding ophthalmology and vision research, extensive interrogation of patient samples for underlying causative DNA mutations has resulted in the discovery of many new genes relevant to eye disease.
Recent Publications
Contact
- nicholas.owen1 [@] gmail.com
- 11-43 Bath St., London, UK, EC1V 9EL
- DM Me
- Zoom Me