Loss of the crumbs cell polarity complex disrupts epigenetic transcriptional control and cell cycle progression in the developing retina
Abstract
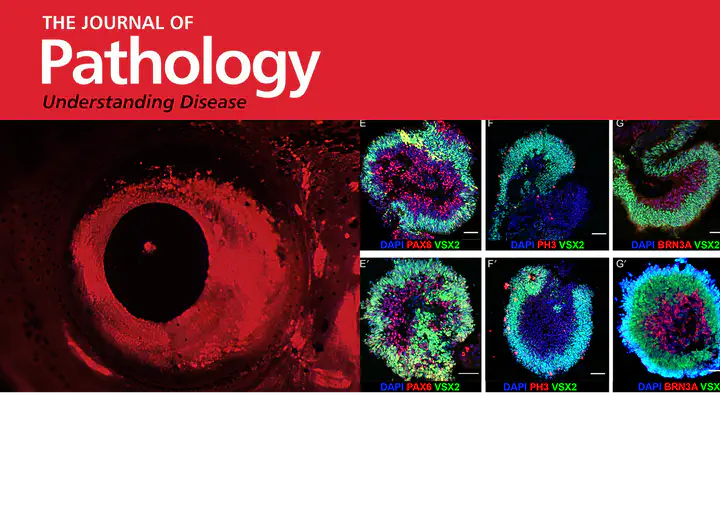
The crumbs cell polarity complex plays a crucial role in apical-basal epithelial polarity, cellular adhesion, and morphogenesis. Homozygous variants in human CRB1 result in autosomal recessive Leber congenital amaurosis (LCA) and retinitis pigmentosa (RP), with no established genotype-phenotype correlation. The associated protein complexes have key functions in developmental pathways; however the underlying disease mechanism remains unclear. Using the oko meduzym289/m289 (crb2a-/-) zebrafish, we performed integrative transcriptomic (RNA-seq data) and methylomic (reduced representation bisulphite sequencing, RRBS) analysis of whole retina to identify dysregulated genes and pathways. Delayed retinal cell specification was identified in both the crb2a-/- zebrafish and CRB1 patient-derived retinal organoids, highlighting dysfunction of cell cycle modulation and epigenetic transcriptional control. Differential DNA methylation analysis revealed novel hypermethylated pathways involving biological adhesion, Hippo and transforming growth factor $\beta$ (TGF$\beta$) signalling. By integrating gene expression with DNA methylation using functional epigenetic modules (FEM), we identified 6 key modules involving cell cycle control and disturbance of TGF$\beta$, BMP, Hippo, and SMAD protein signal transduction pathways, revealing significant interactome hotspots relevant to crb2a function, confirming the epigenetic control of gene regulation in early retinal development and points to a novel mechanism underlying CRB1-retinopathies.